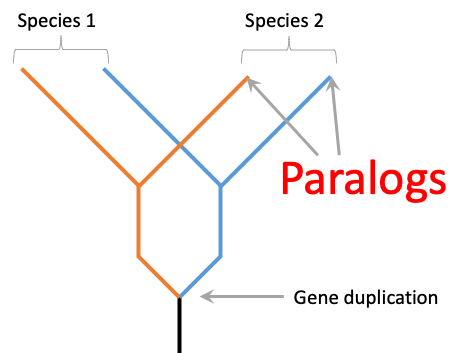
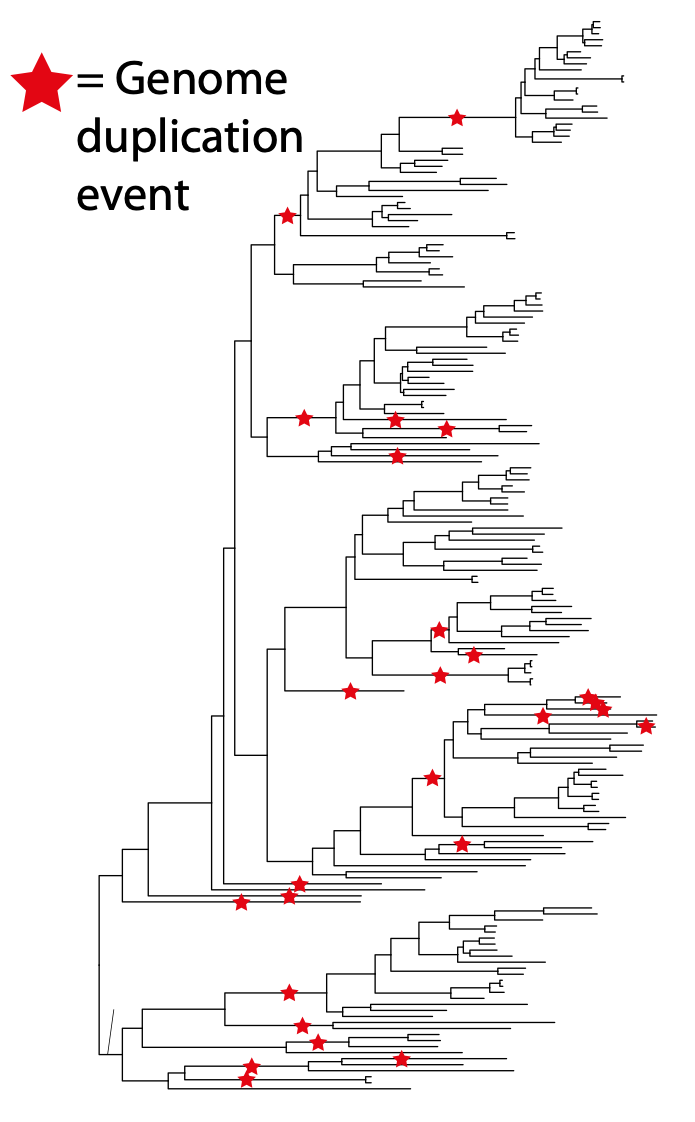
Lab members in bold. Publications with lab members as the first and the last authors are work done mainly in our lab.
Google Scholar
2024 and in progress
Qiu, Y.-J., D. Petrella, F. Sessoms, Y. Yang, M. Esler, C.D. Hirsch, G. Heineck, A. Hegeman, E. Watkins. Evaluating the Effects of Propiconazole on Hard Fescue (Festuca brevipila) via RNA sequencing and Liquid Chromatography–Mass Spectrometry. Preprint
39. Feng, K.-Y., J.F. Walker, H.E. Marx, Y. Yang, S.F. Brockington, M.J. Moore, R.K. Rabeler, S.A. Smith. The link between ancient whole-genome duplications and cold adaptations in the Caryophyllaceae. American Journal of Botany. Open Access
38. Zuntini, A.R., ...279 authors including Y. Yang..., W.J. Baker. Phylogenomics and the rise of the angiosperms. Nature. Open Access.
37. Crum, A.H., L. Philander, L. Busta, Y. Yang. Traditional medicinal use is linked with apparency, not specialized metabolite profiles in the order Caryophyllales. American Journal of Botany. Open Access
36. Pucker, B., N. Walker-Hale, J. Dzurlic, W.C. Yim, J.C. Cushman, A. Crum, Y. Yang, and S.F. Brockington. Multiple mechanisms explain loss of anthocyanins from betalain-pigmented Caryophyllales, including repeated wholesale loss of a key anthocyanidin synthesis enzyme. New Phytologist. 241:471–489. Open Access
Google Scholar
2024 and in progress
Qiu, Y.-J., D. Petrella, F. Sessoms, Y. Yang, M. Esler, C.D. Hirsch, G. Heineck, A. Hegeman, E. Watkins. Evaluating the Effects of Propiconazole on Hard Fescue (Festuca brevipila) via RNA sequencing and Liquid Chromatography–Mass Spectrometry. Preprint
39. Feng, K.-Y., J.F. Walker, H.E. Marx, Y. Yang, S.F. Brockington, M.J. Moore, R.K. Rabeler, S.A. Smith. The link between ancient whole-genome duplications and cold adaptations in the Caryophyllaceae. American Journal of Botany. Open Access
38. Zuntini, A.R., ...279 authors including Y. Yang..., W.J. Baker. Phylogenomics and the rise of the angiosperms. Nature. Open Access.
37. Crum, A.H., L. Philander, L. Busta, Y. Yang. Traditional medicinal use is linked with apparency, not specialized metabolite profiles in the order Caryophyllales. American Journal of Botany. Open Access
- Alex reviewed and categorized traditional medicinal uses across Caryophyllales. Counterintuitively, uses are strongly linked with apparency, not specialized metabolite profiles in the order.
- Still, Alex found weak signals of a positive association between Mental/Nervous System Disorders and Tyr-enriched metabolisms.
- Phylogenetic analysis of traditional medicinal use has many caveats due to the heterogeneity of the data. Alex comprehensively addressed each of them.
36. Pucker, B., N. Walker-Hale, J. Dzurlic, W.C. Yim, J.C. Cushman, A. Crum, Y. Yang, and S.F. Brockington. Multiple mechanisms explain loss of anthocyanins from betalain-pigmented Caryophyllales, including repeated wholesale loss of a key anthocyanidin synthesis enzyme. New Phytologist. 241:471–489. Open Access
|
2023
35. Mohn, R.A. Polyploidy inference across time scales in the charismatic carnivorous plant genus Drosera L. (Droseraceae, Caryophyllales). Ph.D. dissertation. University of Minnesota. 34. Mohn, R.A., R. Zenil-Ferguson, T.A. Krueger, A.S. Fleischmann, A.T. Cross, Y. Yang. Dramatic difference in rate of chromosome number evolution among sundew (Drosera L., Droseraceae) lineages. Evolution. 77(10):2314–2325. Open Access
33. Feng, T., B. Pucker, T.-H. Kuang, B. Song, Y. Yang, N. Lin, H.-J. Zhang, M.J. Moore, S.F. Brockington, Q.-F. Wang, T. Deng, H.-C. Wang & H. Sun. The genome of the glasshouse plant noble rhubarb (Rheum nobile) provides a window into alpine adaptation. Communications Biology, 706 (2023). Open Access |
|
2022
32. Morales-Briones, D.F., N. Lin, E.Y. Huang, D.L. Grossenbacher, J.M. Sobel, C.D. Gilmore, D.C. Tank, Y. Yang. Phylogenomic analyses in Phrymaceae reveal extensive gene tree discordance in relationships among major clades. American Journal of Botany. 109(6): 1035–1064 Open Access
31. Tefarikis, D.T., D.F. Morales-Briones, Y. Yang, G. Edwards, G. Kadereit. On the hybrid origin of the C2 Salsola divaricata agg. (Amaranthaceae) from C3 and C4 parental lineages. New Phytologist. 234:1876–1890. Open access
30. Morales-Briones, D.F., B. Gehrke, C.-H. Huang, A. Liston, H. Ma, H.E. Marx, D.C. Tank, Y. Yang. Analysis of paralogs in target enrichment data pinpoints multiple ancient polyploidy events in Alchemilla s.l. (Rosaceae). Systematic Biology. 71(1):190–207. Open access
|
|
2021
29. Thompson, C.L., M. Alberti, S. Barve, F.U. Battistuzzi, J.L. Drake, G. Casas Goncalves, L. Govaert, C. Partridge, Y. Yang. Back to the future: Reintegrating biology to understand how past eco-evolutionary change can predict future outcomes. Integrative and Comparative Biology. 61(6):2218–2232 Link
28. Shaw, A.K., C. Accolla, J.M. Chacón, T.L. Mueller, M. Vaugeois, Y. Yang, N. Sekar, and D.E. Stanton. Differential retention contributes to racial/ethnic disparity in U.S. academia. PLoS ONE 16(12): e0259710. Open access
27. Qiu, Y., Y. Yang, C.D. Hirsch, E. Watkins. Building a reference transcriptome for the hexaploid hard fescue turfgrass (Festuca brevipila) using a combination of PacBio IsoSeq and Illumina sequencing. Crop Science. 61:2798–2811. Link 26. Carvender-Bares, J., ...22 co-authors... ,Y. Yang. BII-Implementation: The causes and consequences of plant biodiversity across scales in a rapidly changing world. Research Ideas and Outcomes 7:e63850 Open access
25. Morales-Briones, D.F., G. Kadereit, D.T. Tefarikis, M.J. Moore, S.A. Smith, S.F. Brockington, A. Timoneda, W.C. Yim, J.C. Cushman, and Y. Yang. Disentangling sources of gene tree discordance in phylogenomic datasets: Testing ancient hybridizations in Amaranthaceae s.l. Systematic Biology. 70(2):219–235. Open access
|
2020 - Did 2020 ever happen?!
|
2019
24. Qiu, Y.-J., C.D. Hirsch, Y. Yang, and E. Watkins. Towards improved molecular identification tools in Fine Fescue (Festuca L., Poaceae) turfgrasses: Nuclear genome size, ploidy, and chloroplast genome sequencing. Frontiers in Genetics.10:1223. Open access 23. Chen L.-Y., D.F. Morales-Briones, C.N. Passow, and Y. Yang. Performance of gene expression analyses using de novo assembled transcripts in polyploid species. Bioinformatics. 35(21): 4314–4320. Link
22. Feng T., Y. Yang, L. Busta, E.B. Cahoon, H.-C. Wang, and S.-Y. Lü. FAD2 Gene radiation and positive selection contributed to polyacetylene metabolism evolution in Campanulids. Plant Physiology. 181(2):714–728. Open access 21. Wang N., Y. Yang, M.J. Moore, S.F. Brockington, J.F. Walker, J.W. Brown, B. Liang, T. Feng, C. Edwards, J. Mikenas, J. Olivieri, V. Hutchison, A. Timoneda, T. Stoughton, R. Puente, L.C. Majure, U. Eggli, and S.A. Smith. Evolution of Portulacineae marked by gene tree conflict and gene family expansion associated with adaptation to harsh environments. Molecular Biology and Evolution. 36(1): 112–126. Link |
|
2018
20. Yang, Y., C.W. Morden, M.J. Sporck-Koehler, L. Sack, W.L. Wagner, and P.E. Berry. Repeated range expansion and niche shift in a volcanic hotspot archipelago: radiation of C4 Hawaiian Euphorbia (Euphorbiaceae). Ecology and Evolution. doi:10.1002/ece3.4354 Open access
19. Walker, J.F., Y. Yang, T. Feng, A. Timoneda, J. Mikenas, V. Hutchison, C. Edwards, N. Wang, S. Ahluwalia, J. Olivieri, N. Walker-Hale, L.C. Majure, R. Puente, G. Kadereit, M. Lauterbach, U. Eggli, H. Flores-Olvera, H. Ochoterena, S.F. Brockington, M.J. Moore, and S.A. Smith. From cacti to carnivores: Improved phylotranscriptomic sampling and hierarchical homology inference provides further insight to the evolution of Caryophyllales. American Journal of Botany. 105(3):446–462 Open access
18. McKain, M.R., M.G. Johnson, S. Uribe‐Convers, D. Eaton, and Y. Yang. Practical considerations for plant phylogenomics. Applications in Plant Sciences 6(3): e1038. Open access
17. Lopez-Nievesa, S., Y. Yang, A. Timoneda, M.-M. Wang, T. Feng, S.A. Smith, S.F. Brockington, and H.A. Maeda. Relaxation of tyrosine pathway regulation underlies the evolution of betalain pigmentation in Caryophyllales. New Phytologist 217(2): 896–908 Link
16. Yang, Y., M.J. Moore, S.F. Brockington, J. Mikenas, J. Olivieri, J.F. Walker, and S.A. Smith. Improved transcriptome sampling pinpoints 26 ancient and more recent polyploidy events in Caryophyllales, including two allopolyploidy events. New Phytologist. 217(2): 855–870 Open access
15. Smith, S.A., J.W. Brown, Y. Yang, R. Bruenn, C.P. Drummond, S.F. Brockington, J.F. Walker, N. Last, N.A. Douglas, and M.J. Moore. Disparity, diversity, and duplications in the Caryophyllales. New Phytologist 217(2): 836–854 Open access |
|
2017
14. Walker, J.F., Y. Yang, M.J. Moore, J. Mikenas, A. Timoneda, S.F. Brockington, and S.A. Smith. Widespread paleopolyploidy, gene tree conflict, and recalcitrant relationships among the carnivorous Caryophyllales. American Journal of Botany. 104(6): 858–867 Open access 13. Yang, Y., M.J. Moore, S.F. Brockington, A. Timoneda, T. Feng, H. E. Marx, J. F. Walker, and S.A. Smith. An efficient field and laboratory workflow for plant phylotranscriptomic projects. Applications in Plant Sciences 5(3): 1600128. Open access
|
|
2013–2016 (during postdoc in Smith Lab, University of Michigan)
12. Berry, P.E., J.A. Peirson, J.J. Morawetz, V.W. Steinmann, R. Riina, Y. Yang, D. Geltman, and N.I. Cacho. 2016. Euphorbia. Flora of North America Editorial Committee, eds. Flora of North America North of Mexico. Vol. 12. 240–324. New York and Oxford.
11. Smith, S.A., M.J. Moore, J.W. Brown and Y. Yang. 2015. Analysis of phylogenomic datasets reveals conflict, concordance, and gene duplications with examples from animals and plants. BMC Evolutionary Biology. 15:150. doi:10.1186/s12862-015-0423-0 Open access
10. Brockington S.F.*, Y. Yang*, F. Gandia-Herrero, S. Covshoff, J.M. Hibberd, R.F. Sage, G.K.-S. Wong, M.J. Moore and S.A. Smith. 2015. Lineage specific gene radiations underly the evolution of novel betalain pigmentation in Caryophyllales. New Phytologist. 207(4) 1170–1180 *Co-first author. Open access Commentary by David L. Des Marais
9. Yang, Y., M.J. Moore, S.F. Brockington, D.E. Soltis, G.K.-S. Wong, E.J. Carpenter, Y. Zhang, L. Chen, Z.-X. Yan, Y.-L. Xie, R.F. Sage, S. Covshoff, J.M. Hibberd, M.N. Nelson, and S.A. Smith. 2015. Dissecting molecular evolution in the highly diverse plant clade Caryophyllales using transcriptome sequencing. Molecular Biology and Evolution. 32(8): 2001–2014. Open access
8. Horn, J.W., Z.-X. Xi, R. Riina, J.A. Peirson, Y. Yang, B.L. Dorsey, P.E. Berry, C.C. Davis, and K.J. Wurdack. 2014. Evolutionary bursts in the Euphorbia (Euphorbiaceae) are linked with photosynthetic pathway. Evolution. 68(12): 3485–3504 Link
7. Yang, Y. and S.A. Smith. 2014. Orthology inference in non-model organisms using transcriptomes and low-coverage genomes: improving accuracy and matrix occupancy for phylogenomics. Molecular Biology and Evolution. 31 (11): 3081–3092 Open access
6. Yang, X.-X, Y. Yang, C.-J. Ji, T. Feng, Y. Shi, L. Lin, J.-J. Ma, & J.-S. He. 2014. Large-scale patterns of stomatal traits in Tibetan and Mongolian grassland species. Basic and Applied Ecology 15(2):122–132 Link
5. Yang, Y. and S.A. Smith. 2013. Optimizing de novo assembly of short-read RNA-seq data for phylogenomics. BMC Genomics 14:328. Open access
|
|
2012 and earlier (during graduate school in Berry Lab, University of Michigan)
4. Yang, Y., R. Riina, J.J. Morawetz, T. Haevermans, X. Aubriot and P.E. Berry. 2012. Molecular phylogenetics and classification of Euphorbia subgenus Chamaesyce: a group with high diversity in photosynthetic systems and growth forms. Taxon 61(4):764–789 Link
3. Yang, Y. and P.E. Berry. 2011. Phylogenetics of the Chamaesyce clade (Euphorbia, Euphorbiaceae): reticulate evolution and long-distance dispersal in a prominent C4 lineage. American Journal of Botany 98(9):1486–1503 Link
2. Berry, P. E., V. Steinmann, and Y. Yang. 2011. Proposal to conserve the name Euphorbia acuta Engelm. against Euphorbia acuta Bellardi ex Colla (Euphorbiaceae). Taxon 60(2): 603–604 Link 1. Hendry, T.A., B. Wang, Y. Yang, E.C. Davis, J.E. Braggins, R.M. Schuster, & Y.-L. Qiu.2007. Evaluating phylogenetic positions of four liverworts from New Zealand, Neogrollea notabilis, Jackiella curvata, Goebelobryum unguiculatum and Herzogianthus vaginatus, using three chloroplast genes. The Bryologist 110: 738–751 Link
|